 |
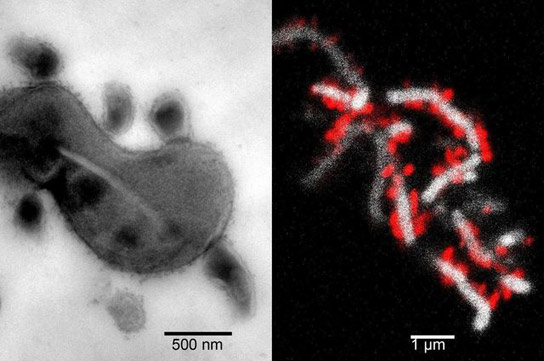
| At left, the tight physical association between TM7x cells and XH001. At right, TM7x cells (red) attach to the surface of XH001 (white) | sci-english.blogspot.com |
One of the great recent discoveries in modern biology was that the human body contains 10 times more bacterial cells than human cells. But much of that bacteria is still a puzzle to scientists.
It is estimated by scientists that roughly half of bacteria living in human bodies is difficult to replicate for scientific research — which is why biologists call it “microbial dark matter.” Scientists, however, have long been determined to learn more about these uncultivable bacteria, because they may contribute to the development of certain debilitating and chronic diseases.
For decades, one bacteria group that has posed a particular challenge for researchers is the Candidate Phylum TM7, which has been thought to cause inflammatory mucosal diseases because it is so prevalent in people with periodontitis, an infection of the gums.
Now, a landmark discovery by scientists at the UCLA School of Dentistry, the J. Craig Venter Institute and the University of Washington School of Dentistry has revealed insights into TM7’s resistance to scientific study and to its role in the progression of periodontitis and other diseases. Their findings shed new light on the biological, ecological and medical importance of TM7, and could lead to better understanding of other elusive bacteria.
The team’s findings are published online in the December issue of the Proceedings of the National Academy of Sciences.
“I consider this the most exciting discovery in my 30-year career,” said Dr. Wenyuan Shi, a UCLA professor of oral biology. “This study provides the roadmap for us to make every uncultivable bacterium cultivable.”
The researchers cultivated a specific type of TM7 called TM7x, a version of TM7 found in people’s mouths, and found the first known proof of a signaling interaction between the bacterium and an infectious agent called Actinomyces odontolyticus, or XH001, which causes mucosal inflammation.
“Once the team grew and sequenced TM7x, we could finally piece together how it makes a living in the human body,” said Dr. Jeff McLean, acting associate professor at the University of Washington School of Dentistry. “This may be the first example of a parasitic long-term attachment between two different bacteria — where one species lives on the surface of another species gaining essential nutrients and then decides to thank its host by attacking it.”
To prove that TM7x needs XH001 to grow and survive, the team attempted to mix isolated TM7x cells with other strains of bacteria. Only XH001 was able to establish a physical association with TM7x, which led researchers to believe that TM7x and XH001 might have evolved together during their establishment in the mouth.
What makes TM7x even more intriguing are its potential roles in chronic inflammation of the digestive tract, vaginal diseases and periodontitis. The co-cultures collected in this study allowed researchers to examine, for the first time ever, the degree to which TM7x helps cause these conditions.
“Uncultivable bacteria presents a fascinating ‘final frontier’ for dental microbiologists and are a high priority for the NIDCR research portfolio,” said Dr. R. Dwayne Lunsford, director of the National Institute of Dental and Craniofacial Research’s microbiology program. “This study provides a near-perfect case of how co-cultivation strategies and a thorough appreciation for interspecies signaling can facilitate the recovery of these elusive organisms. Although culture-independent studies can give us a snapshot of microbial diversity at a particular site, in order to truly understand physiology and virulence of an isolate, we must ultimately be able to grow and manipulate these bacteria in the lab.”
It was previously known that XH001 induces inflammation. But by infecting bone marrow cells with XH001 alone and then with the TM7x/XH001 co-culture, the researchers also found that inflammation was greatly reduced when TM7x was physically attached to XH001. This is the only known study that has provided evidence of this relationship between TM7 and XH001.
The researchers plan to further study the unique relationship between TM7X and XH001 and how they jointly cause mucosal disease. Their findings could have implications for potential treatment and therapeutics.
Other collaborators on the study were Drs. Xuesong He, Renate Lux and Anna Edlund of the UCLA School of Dentistry; Shibu Yooseph, Adam Hall and Karen Nelson of the Venter Institute; Su-Yang Liud and Genhong Cheng of the UCLA department of microbiology, immunology and molecular genetics; Pieter Dorresteine of UC, San Diego; Eduardo Esquenazi of Sirenas Marine Discovery; and Ryan Hunter of the University of Minnesota.
Dr. Shi is part-time chief science officer at C3 Jian, Inc., which has licensed technologies from the University of California Regents that could be indirectly related to this research project.
The work was supported in part by the NIH’s National Institute of Dental and Craniofacial Research (grants 1R01DE023810-01, 1R01DE020102 and 1R01DE021108, and T90 training award DE022734) and the National Institute of General Medical Sciences (grant 1R01GM095373).
Publication: Xuesong He, et al., “Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle,” PNAS, 2014; doi: 10.1073/pnas.1419038112
Source: Brianna Aldrich, UCLA News
Image: Batbileg Bor/UCLA and Ryan Hunter/University of Minnesota
No comments:
Post a Comment